I liked the question and it motivated me to find a solution.
I have tried everything and this gives the best result with the least effort:
You need the “File Actions app” files_scripts (build the lua php-module as described → here ← )
Then you need pandoc and soffice (Libre Office) on the host system.
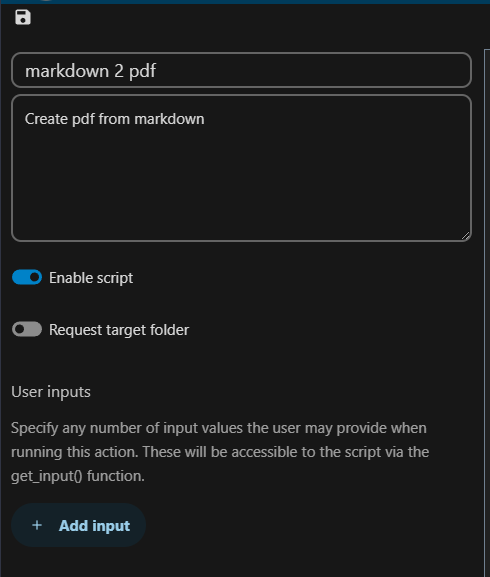
Create a “New action” in the admin section of “File actions”, give it a name like “markdown 2 pdf”:
and paste this code-block into the code-window:
local files = get_input_files()
-- finding out nextclouds installation dir with shell_command `pwd`
local current_dir = shell_command('pwd').output
-- Loop through input files
for _, input_file in ipairs(files) do
local log_filename = input_file.name:gsub('%.[Mm][DdWwNnTtXx]+$', '.pdf') .. '.encoding.txt'
local outfolder = get_parent(input_file)
local out_meta = meta_data(outfolder)
-- Check if the logfile already exists
if exists(outfolder, log_filename) then
abort('Logfile "' .. outfolder.path .. "/" .. outfolder.name .. "/" .. log_filename .. '" already exists. Skipping...')
goto continue
end
local arguments = {}
if is_file(input_file) then
local html_filename = input_file.name:gsub('%.[Mm][DdWwNnTtXx]+$', '.html')
local out_filename = input_file.name:gsub('%.[Mm][DdWwNnTtXx]+$', '.pdf')
-- Check if the output file already exists
if exists(outfolder, out_filename) then
abort('A file "' .. out_filename .. '" already exists in the target directory. Skipping...')
goto continue
end
-- meta_data part:
local in_meta = meta_data(input_file)
local metadata_args = {
-- obtained from shell_command('pwd')
"\"nextcloud_dir = " .. current_dir .. "\"",
"\"in_file = " .. input_file.name .. "\"",
"\"in_mimetype = " .. in_meta.mimetype .. "\"",
"\"local_path = " .. out_meta.local_path .. "\"",
"\"html_filename = " .. html_filename .. "\"",
"\"out_filename = " .. out_filename .. "\"",
"\"out_storage_path = " .. out_meta.storage_path .. "/" .. out_filename .. "\"",
}
-- Concatenate the metadata_args array into a single string with spaces between each argument
local arguments_string = table.concat(metadata_args, " ")
table.insert(arguments, arguments_string)
end
-- Construct the command string with quotes around each argument
local command = "/usr/local/bin/nc-md-to-pdf " .. table.concat(arguments, " ")
-- Run the command
local result = shell_command(command)
-- Create debug file with the same name as the output file but with extra .encoding.txt extension
local output_file = new_file(outfolder, log_filename, result.output .. "\n" .. result.errors)
if output_file == nil then
abort("Could not create logfile: " .. log_filename)
end
::continue::
end
Activate and store it.
Now create a bash-script
/usr/local/bin/nc-md-to-pdf
with this content:
#!/bin/bash
echo "$(date +"%Y-%m-%d %H:%M:%S %Z")"
if [ $# -lt 10 ]; then digits=1
elif [ $# -lt 100 ]; then digits=2
else digits=3
fi
# iterate over the arguments
for (( i=0; i<=$#; i++ )); do
# format the index with leading zeros
index=$(printf "%0*d" $digits $i)
# get the i-th argument
arg=${!i}
# echo the argument with its index
echo " - arg $index = $arg"
echo "$arg" | grep -q "nextcloud_dir" && NCC="${arg##*= }/occ"
# in:
echo "$arg" | grep -q "in_file" && in_file="${arg##*= }"
echo "$arg" | grep -q "in_mimetype" && in_mt="${arg##*= }"
echo "$arg" | grep -q "local_path" && local_path="${arg##*= }"
# out:
echo "$arg" | grep -q "html_filename" && html_fn="${arg##*= }"
echo "$arg" | grep -q "out_filename" && out_fn="${arg##*= }"
echo "$arg" | grep -q "out_storage_path" && out_sp="${arg##*= /}"
done
echo
in_lp="$local_path/$in_file"
mime_type=$(file -bL --mime-type "$in_lp")
echo "information gathered by file:"
echo " file = $(file -bL "$in_lp")"
echo " --mime-type = $mime_type"
echo "NCC = $NCC"
echo "in_file = $in_file"
echo "in_mt = $in_mt"
echo "local_path = $local_path"
echo "html_fn = $html_fn"
echo "out_fn = $out_fn"
echo "out_sp = $out_sp"
#exit 0
# cd into infile dir to be as relative to images as the md file
cd "$local_path"
if [ "$in_mt" = "text/markdown" ]; then
if pandoc -f markdown -t html "$in_file" -o "$html_fn" && soffice --headless --convert-to pdf "$html_fn"; then
rm "$html_fn"
$NCC files:scan --verbose --no-interaction --path="$out_sp"
fi
else
echo "$in_file seems not to be a markdown file, skipping"
fi
exit 0
(last updated 2023.03.24 12:50 CET)
and make it executable:
chmod +x /usr/local/bin/nc-md-to-pdf
It creates the pdf file and a log file for debug purposes.
This is a quick sketch and can be fleshed out as you like. So did I not yet work on image resize templates.
You can trigger it from the context menu but it is possible to make a background job with the flow mechanism, as described here